Critical Assessment of protein Structure Prediction (CASP)
The Critical Assessment of protein Structure Prediction (CASP) is a community-wide prediction competition where research groups are required to predict 3-D structures from protein sequences.
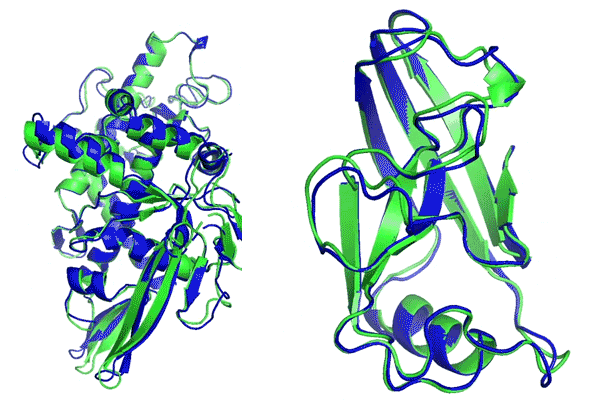
More than 100 research groups worldwide join the CASP competition every two years. Using all sequence and structure data available at present time, they predict structures for targets with newly derived (yet unreleased) structures, specifically withheld from the public. Targets are divided into easier (template-based) and harder (template-free, de novo) categories.
The main evaluation metric used is the Global Distance Test – Total Score (GDT-TS). It measures what percentage of α-carbons in the predicted structure are within a threshold distance (1, 2, 4, 8 Å) of the known structure, for the best possible alignment of the two.
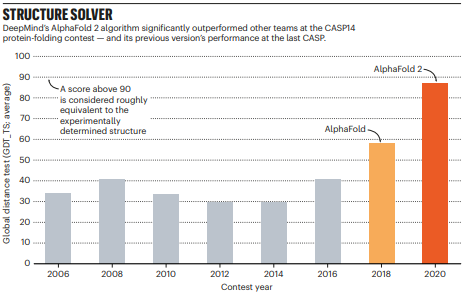
After years of stagnation, DeepMind entered the competition for the first time in 2018, and won with a significant lead on the other contenders. In 2020, the prediction results of their updated system, AlphaFold 2.0, were so good that the structure prediction problem has been dubbed as solved by some.